01 Introduction to SpaDES
Alex M. Chubaty
Eliot J. B. McIntire
September 18 2025
Source:vignettes/i-introduction.Rmd
i-introduction.RmdIntroduction to SpaDES
Package description
Easily implement a variety of simulation models, with a focus on spatially explicit models. These include raster-based, event-based, and agent-based models. The core simulation components are built upon a discrete event simulation framework that facilitates modularity, and easily enables the user to include additional functionality by running user-built simulation modules. Included are numerous tools to rapidly visualize raster and other maps.
Objectives and motivations
Building spatial simulation models often involves reusing various model components, often having to re-implement similar functionality in multiple simulation frameworks (i.e, in different programming languages). When various components of a simulation model become fragmented across multiple platforms, it becomes increasingly difficult to link these various components, and often solutions for this problem are idiosyncratic and specific to the model being implemented. As a result, developing general insights into complex computational models has, in the field of ecology at least, been hampered by modellers’ typically developing models from scratch (Thiele and Grimm 2015).
SpaDES is a generic simulation platform that can be used
to create new model components quickly. It also provides a framework to
link with existing simulation models, so that an already well described
and mature model, e.g., Landis-II (Scheller et al. 2007), can be used with de
novo components. Alternatively one could use several de
novo models and several existing models in combination. This
approach requires a platform that allows for modular reuse of model
components (herein called ‘modules’) as hypotheses that can be evaluated
and tested in various ways, as advocated by Thiele and Grimm (2015).
When beginning development of this package, we sought a general simulation platform at least the following characteristics:
- Allow rapid building of models of a wide diversity of types (e.g., agent-based models, raster models, differential equation models);
- Run faster and more memory efficiently than current systems for doing similar things (e.g., NetLogo (Wilensky 1999), SELES (Fall and Fall 2001));
- Use a platform that already has strong data analysis, manipulation, and visualization capacities;
- Be open source, but also make it as easy as possible for many people to easily contribute modules and code;
- Be easy to use for a large number of scientists who aren’t formally trained as computer programmers or have limited programming experience;
- Should be built around modularity so that models can be seen as modules that are easily replaceable, not just ‘in theory’ replaceable;
- Allow tight coupling between data and model simulations so that the entire work flow from raw data to result generation or even report generation is not actually something that one has to redesign every time there is a new data set.
We selected R as the system within which to build
SpaDES. R is currently the lingua
franca for scientific data analysis. This means that anything
developed in SpaDES is simply R code and can
be easily shared with journals and the scientific community. We can
likewise leverage R’s strengths as a data platform, its
excellent visualization and graphics, its capabilities to run external
code such as C/C++ and easily interact
external software such as databases, and its abilities for high
performance computing. SpaDES therefore doesn’t need to
implement all of these from scratch, as they are achievable with already
existing R packages.
Is R fast enough?
High-level programming languages are often criticized for being much
slower than their low-level counterparts. R has definitely
received its share of criticism over not just speed but also memory
use1.
While some of these criticisms may be legitimate, many of them are
overblown. They are based on test code that is not written for how
R works[^benchmarks]. The best example of this is using
traditional C-like loops in R. R
is a vectorized language, and code should rarely be written as loops in
R. It should be vectorized, yet these sorts of biased
comparisons get made all of the time, giving the false impression that
R is too slow. Thus, these tests show that poorly written
code can be slow, in any language.
Another major criticism of R is its high memory
footprint. It’s true that similar structures take up less memory in
C than in R. However, there are various simple
optimizations for R code, such as explicitly pre-allocating
memory to objects, that can drastically improve performance. In cases
were further improvements are required, the Rcpp package
(Eddelbuettel and Francois 2011; Eddelbuettel
2013) allows easy writing of low memory footprint
C++ code that can then be called in R.
Likewise, numerous upgrades to R, including minimizing object copying
since R version 3.0, and extremely powerful
user developed packages, like data.table and
dplyr are eliminating many of these concerns.
Discrete event simulation and SpaDES
Discrete event simulation (DES) as implemented here is ‘event driven’, meaning that an activity changes the state of the system at particular times (called events). This approach assumes that state of the system only changes due to events, therefore there is no change between events. A particular activity may have several events associated with it. Future events are scheduled in an event queue, and then processed in chronological order (with ties being resolved using ‘first-in-first-out’). Because the system state doesn’t change between events, we do not need to ‘run the clock’ in fixed increments each timestep. Rather, time advances to the time of the next event in the queue, effectively optimizing computations especially when different modules have different characteristic time intervals (i.e., ‘timestep’).
‘Time’ is the core concept linking various simulation components via
the event queue. Activities schedule events (which change the state of
the system according to their programmed rules) and do not need to know
about each other. Rather than wrapping a sequence of functions (events)
inside a for loop for time and iterating through each
timestep, each event is simply scheduled to be completed. Repeated
events are simply scheduled repeatedly. This not only allows for
modularity of simulation components, it also allows complex model
dynamics to emerge based on scheduling rules of each activity (module).
Thus, complex simulations involving multiple processes (activities) can
be built fairly easily, provided these processes are modelled using a
common DES framework.
SpaDES provides such a framework, facilitating
interaction between multiple processes (built as ‘modules’) that don’t
interact with one another directly, but are scheduled in the event queue
and carry out operations on shared data objects in the simulation
environment. This package provides tools for building modules natively
in R that can be reused. Additionally, because of the
flexibility R provides for interacting with other
programming languages and external data sources, modules can also be
built using external tools and integrated with SpaDES (see
figure below).
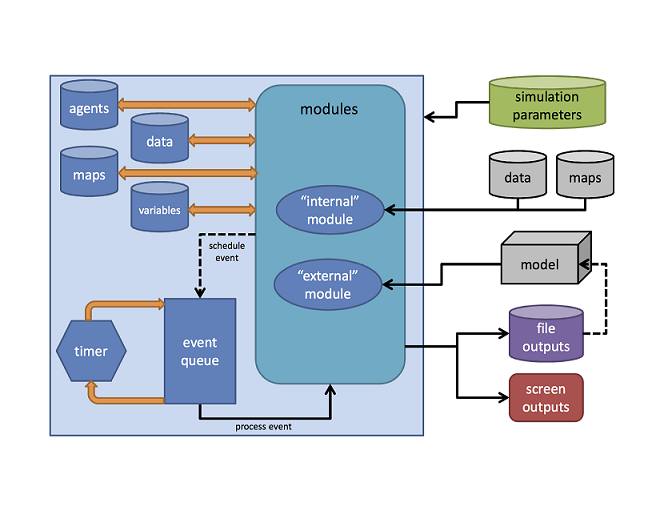
SpaDES modules
A SpaDES module describes the processes or activities
that drive simulation state changes via changes to objects stored in the
simulation environment. Each activity consists of a collection of events
which are scheduled depending on the rules of the simulation. Each event
may evaluate or modify a simulation data object (e.g., update
the values on a raster map), or perform other operations such as saving
and loading data objects or plotting.
The power of SpaDES is in modularity and the relative
ease with which existing modules can be modified and new modules
created, in native R as well as through the incorporation
of external simulation modules. Creating and customizing modules is a
whole topic unto itself, and for that reason we have created a separate
modules vignette with more details on
module development.
Strict modularity requires that modules can act independently,
without needing to know about other modules. However, what if two (or
more) modules are incompatible with one another? To address this, each
SpaDES module is required to explicitly state its input
dependencies (data, package, and parameterization requirements), data
outputs, as well as provide other useful metadata and documentation for
the user. Upon initialization of a simulation, the dependencies of every
module used are examined and evaluated. If dependency incompatibilities
exists, the initialization fails and the user is notified.
SpaDES demos and sample modules
The PDF format does not allow us to demonstrate the simulation visualization components of this package, so we invite you to run the sample simulation provided in this vignette, and to view the source code for the sample modules included in this package.
This demo loads three sample modules provided with the packages: 1)
randomLandscapes, 2) fireSpread, and 3)
caribouMovement. These sample modules, respectively,
highlight several keys features of the package: 1) the import, update,
and plotting of raster map layers; 2) the computational speed of
modeling spatial spread processes; and 3) the implementation of an
agent-based (a.k.a., individual-based) model.
## NOTE: Suggested packages SpaDES.tools and NLMR packages must be installed
#install.packages("SpaDES.taols")
#install.packages("NLMR", repos = "https://predictiveecology.r-universe.dev/")
knitr::opts_chunk$set(eval = requireNamespace("SpaDES.tools") && !requireNamespace("NLMR"))
library(SpaDES.core)
demoSim <- suppressMessages(simInit(
times = list(start = 0, end = 100),
modules = "SpaDES_sampleModules",
params = list(
.globals = list(burnStats = "nPixelsBurned"),
randomLandscapes = list(
nx = 1e2, ny = 1e2, .saveObjects = "landscape",
.plotInitialTime = NA, .plotInterval = NA, inRAM = TRUE
),
caribouMovement = list(
N = 1e2, .saveObjects = "caribou",
.plotInitialTime = 1, .plotInterval = 1, moveInterval = 1
),
fireSpread = list(
nFires = 1e1, spreadprob = 0.235, persistprob = 0, its = 1e6,
returnInterval = 10, startTime = 0,
.plotInitialTime = 0, .plotInterval = 10
)
),
path = list(modulePath = getSampleModules(tempdir()))
))
spades(demoSim)Additional SpaDES modules are available via a GitHub
repository: https://github.com/PredictiveEcology/SpaDES-modules.
Modules from this repository can be downloaded to a local directory
using:
downloadModule(name = "moduleName")Note: by default, modules and their data are saved
to the directory specified by the spades.modulesPath. An
alternate path can be provided to downloadModule directly
via the path argument, or specified using
options(spades.modulesPath = "path/to/my/modules").
A detailed guide to module development is provided in the modules vignette.
Simulation and data
Historically, simulation models were built separately from the analysis of input data (e.g., via regression) and outputs of data (e.g., graphically, statistically). On the input data side, this effectively broke the linkage between data (e.g., from field or satellites) and the simulation. This has the undesired effect of creating the appearance of reduced uncertainty in simulation model predictions, by breaking correlations between parameter estimates (that invariably occur in analyses of real data), or simply by passing an incorrectly specified parameter uncertainty to a simulation model.
Conversely, on the data output side, numerous tools, such as
optimization (e.g., pattern oriented
modeling Grimm et al. 2005) or statistical analyses could not
directly interact with the simulation model, unless a specific extension
was built for that purpose. In R, those tools already exist
and are robust. Thus, validation, calibration, and verification of
simulation models can become rolled into the simulation model itself,
facilitating understanding of models’ forecasting performance and thus
their predictive capacity. All of these enhance transparency and
reproducibility, both desired properties for scientific studies.
Linking the raw data, data analysis, validation, calibration (via optimization), simulation forecasting, and output analyses into a single work flow allow for several powerful outcomes:
- updates to raw data can be easily propagated into the outputs;
- uncertainty in raw data can be passed through to simulations;
- decision makers can be given tools that draw directly on raw data and so lag times between data updates and decision making updates can be dramatically shortened;
- feedbacks between disciplinary silos can be simulated, promoting disciplinary integration.
Using SpaDES
As you can see in the sample simulation code provided above, setting
up and running a simulation in SpaDES is straightforward
using existing modules. You need to specify some things about the
simulation environment including 1) which modules to use for the
simulation, and 2) any data objects (e.g., parameter values)
that should be used to store the simulation state. Each of these are
passed as named lists to the simulation object upon initialization.
Initializing a simulation: the simInit function
The details of each simulation are stored in a simList
object, including the simulation parameters and modules used, as well as
storing the current state of the simulation and the future event queue.
A list of completed events is also stored, which can provide useful
debugging information. This simList object contains a
unique unique object, along with data used/created during
the simulation. The envir object is simply an environment,
which means objects stored in it are updated using reference semantics
(so objects don’t need to be copied). You can access objects stored in
environments using the same syntax as for lists (e.g.,
$, [[), in addition to get, which
makes working with simulated data easy to do inside modules.
A new simulation is initialized using the simInit
function, which does all the work of creating the envir and
simList objects for your simulation. This function also
attempts to provide additional feedback to the user regarding parameters
that may be improperly specified.
Once a simulation is initialized you can inspect the contents of a
envir object using:
# full simulation details:
# simList object info + simulation data
mySim
# less detail:
# simList object isn't shown; object details are
ls.str(mySim)
# least detail:
# simList object isn't shown; object names only
ls(mySim)Simulation module object dependencies can be viewed using:
library(igraph)
library(DiagrammeR)
depsEdgeList(mySim, FALSE) # data.frame of all object dependencies
moduleDiagram(mySim) # plots simplified module (object) dependency graph
objectDiagram(mySim) # plots object dependency diagramThey can viewed directly by printing the output of the
depsEdgeList function.
Running a simulation: the spades function
Once a simulation is properly initialized it is executed using the
spades function. By default in an interactive session, a
progress bar is displayed in the console (this can be customized), and
any specified files are loaded (via including an input
data.frame or data.table, see examples).
Debugging mode, i.e., setting
spades(mySim, debug = TRUE), prints the contents of the
simList object after the completion of every event during
simulation. See the wiki
entry on debugging for more details on debugging SpaDES models.
options(spades.nCompleted = 50) # default: store 1000 events in the completed event list
mySim <- simInit(...) # initialize a simulation using valid parameters
mySim <- spades(mySim) # run the simulation, returning the completed sim object
eventDiagram(mySim) # visualize the sequence of events for all modulesTypes of models
SpaDES provides a common platform for simulation model
development and analysis. As such, its possible to implement and
integrate a wide variety of model types as modules in
SpaDES, for example:
- cellular automata;
- matrix transition models;
- population and metapopulation dynamics;
- agent based (individual based) models;
- GIS/spatial analyses;
- and many others.
The common denominator is the idea of an event. If an event can be
scheduled, i.e., it can be conceived of as having a ‘time’ at
which it occurs, then it can be used with SpaDES. This, of
course, includes static elements that occur only once, such as a the
start of a simulation.
Modelling spread processes
Spatially explicit modules will sometimes contain ‘contagious’
processes, such as spreading (e.g., fires), dispersal
(e.g., seeds), flow (e.g., water or wind). At the core
of SpaDES are a few functions to do these that are
relatively fast computationally. More contagious processes are being
actively being developed.
A simple fire model
Using the spread function, we can simulate fires, and
subsequent changes to the various map layers. Here,
spreadProb can be a single probability or a raster map
where each pixel has a probability. In the demo below, each cell’s
probability is taken from the percentPine map layer.
Agent based models
A primary goal of developing SpaDES was to facilitate
the development of agent-based models (ABMs), also known as
individual-based models (IBMs).
Point agents
As ecologists, we are often concerned with modelling individuals
(agents) in time and space, and whose spatial location (position) can be
represented as a single point on a map. These types of agents can easily
be represented most simply by a single set of coordinates indicating
their current position, and can simulated using a
SpatialPoints object. Additionally, a
SpatialPointsDataFrame can be used, which provides storage
of additional information beyond agents’ coordinates as needed.
Polygon agents
Analogously, it is possible to use SpatialPolygons*.
Plotting methods using Plot are optimized for speed and are
much faster than the default plot methods for polygons (via
spatial subsampling of vector data), so have fewer options for
customization than other approaches to visualizing polygons.
Simulation experiments
Running multiple simulations with different parameter values is a
critical part of sensitivity and robustness analysis, simulation
experiments, optimization, and pattern oriented modelling. Likewise,
greater understanding and evaluation of models and their uncertainty
requires simulation replication and repetition (Grimm and Railsback 2005; Thiele and Grimm
2015). Using R as a common platform for data,
simulation, and analyses we can do all of these easily and directly as
part of our SpaDES simulation without breaking the linkages
between model and analysis. This workflow facilitates and enhances the
use of ensemble and consensus modelling, and studies of cumulative
effects. The tools for these experiments are now in a separate package,
SpaDES.experiment.
Additional resources
SpaDES documentation and vignettes
Help files
From within R, typing ?'spades-package',
will give an categorized view of the functions within
SpaDES.
Vignettes
The following package vignettes are intended to be read in sequence,
and follow a progression from higher-level package organization and
motivation, to detailed implementation of user-built modules and
simulations. To view available vignettes use
browseVignettes(package = "SpaDES.core") (this will only be
available from a CRAN download).
SpaDES module repository
We provide a number of modules to facilitate getting started:
https://github.com/PredictiveEcology/SpaDES-modules
Modules from this (or another suitable GitHub repository) can be
downloaded using downloadModule.
We welcome additional contributions to this module repository.
Reporting bugs
As with any software, there are likely to be issues. If you believe you have found a bug, please contact us via the package GitHub site: https://github.com/PredictiveEcology/SpaDES/issues. Please do not use the issue tracker for general help requests.
For general help with SpaDES and module development
please see our Q&A Forum https://groups.google.com/d/forum/spades-users.